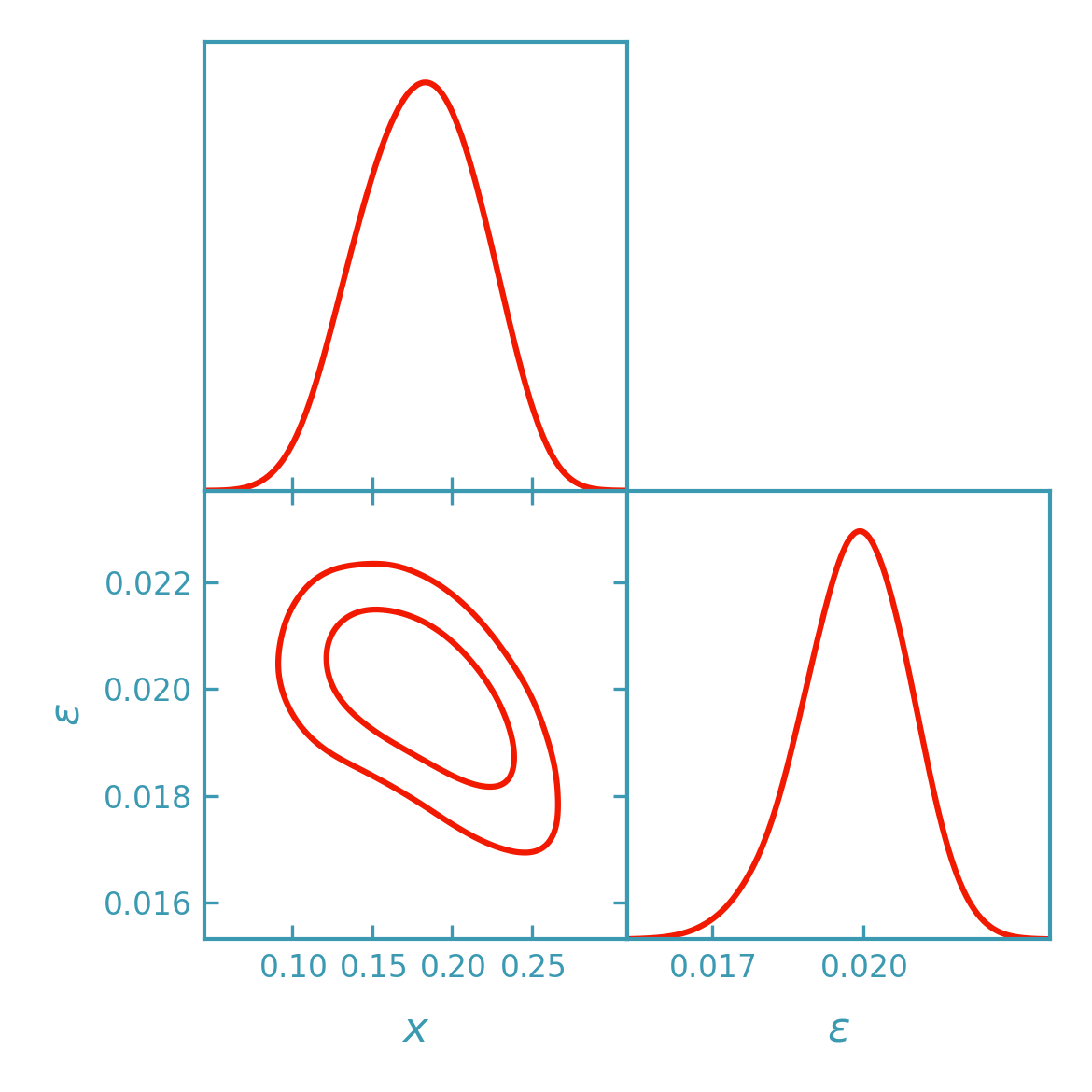
 DALI contours#
DALI contours#
This page shows how to visualize DALI-expanded posteriors using GetDist,
starting from DALI tensors (Fisher F and higher-order DALI tensors).
The focus here is what to do next once you already have a DALI expansion: how to draw samples for inspection, comparison, and posterior analysis.
All posterior quantities shown here refer to the DALI-expanded approximation.
If you are looking for:
how DALI tensors are defined and interpreted, see ForecastKit
how to compute DALI tensors with DerivKit, see DALI tensors
Two workflows are supported:
sampling the DALI log-posterior (
emcee)fast importance sampling using a Fisher–Gaussian proposal
Both workflows return a getdist.MCSamples object.
For a conceptual overview of DALI forecasting, its interpretation, and other forecasting frameworks in DerivKit see ForecastKit.
Sampling the DALI posterior with emcee#
>>> import numpy as np
>>> from getdist import plots as getdist_plots
>>> from derivkit import ForecastKit
>>> def model_2d(theta):
... # Nonlinear forward model with a curved parameter degeneracy
... # (informally referred to as a "banana"-shaped posterior).
... x, eps = float(theta[0]), float(theta[1])
... k = 3.0
... a = 4.0
... c = 6.0
... o1 = 1e2 * np.exp((x - k * eps) ** 2) * np.exp(a * eps)
... o2 = 4e1 * np.exp(0.5 * x) * (1.0 + 0.3 * eps + c * (eps**3))
... return np.array([o1, o2], dtype=float)
>>> theta0 = np.array([0.18, 0.02], dtype=float)
>>> cov = np.array([[1.0, 0.95],
... [0.95, 1.0]], dtype=float)
>>> fk = ForecastKit(function=model_2d, theta0=theta0, cov=cov)
>>> dali = fk.dali(forecast_order=2)
>>> F = dali[1][0]
>>> D1, D2 = dali[2]
>>> samples = fk.getdist_dali_emcee(
... dali=dali,
... names=["x", "eps"],
... labels=[r"x", r"\epsilon"],
... label="DALI (emcee)",
... )
>>> dk_red = "#f21901"
>>> dk_yellow = "#e1af00"
>>> line_width = 1.5
>>> plotter = getdist_plots.get_subplot_plotter(width_inch=3.9)
>>> plotter.settings.linewidth_contour = line_width
>>> plotter.settings.linewidth = line_width
>>> plotter.settings.figure_legend_frame = False
>>> plotter.settings.legend_rect_border = False
>>> plotter.triangle_plot(
... [samples],
... params=["x", "eps"],
... filled=False,
... contour_colors=[dk_red],
... contour_lws=[line_width],
... contour_ls=["-"],
... )
>>> samples.numrows > 0
True
(png)
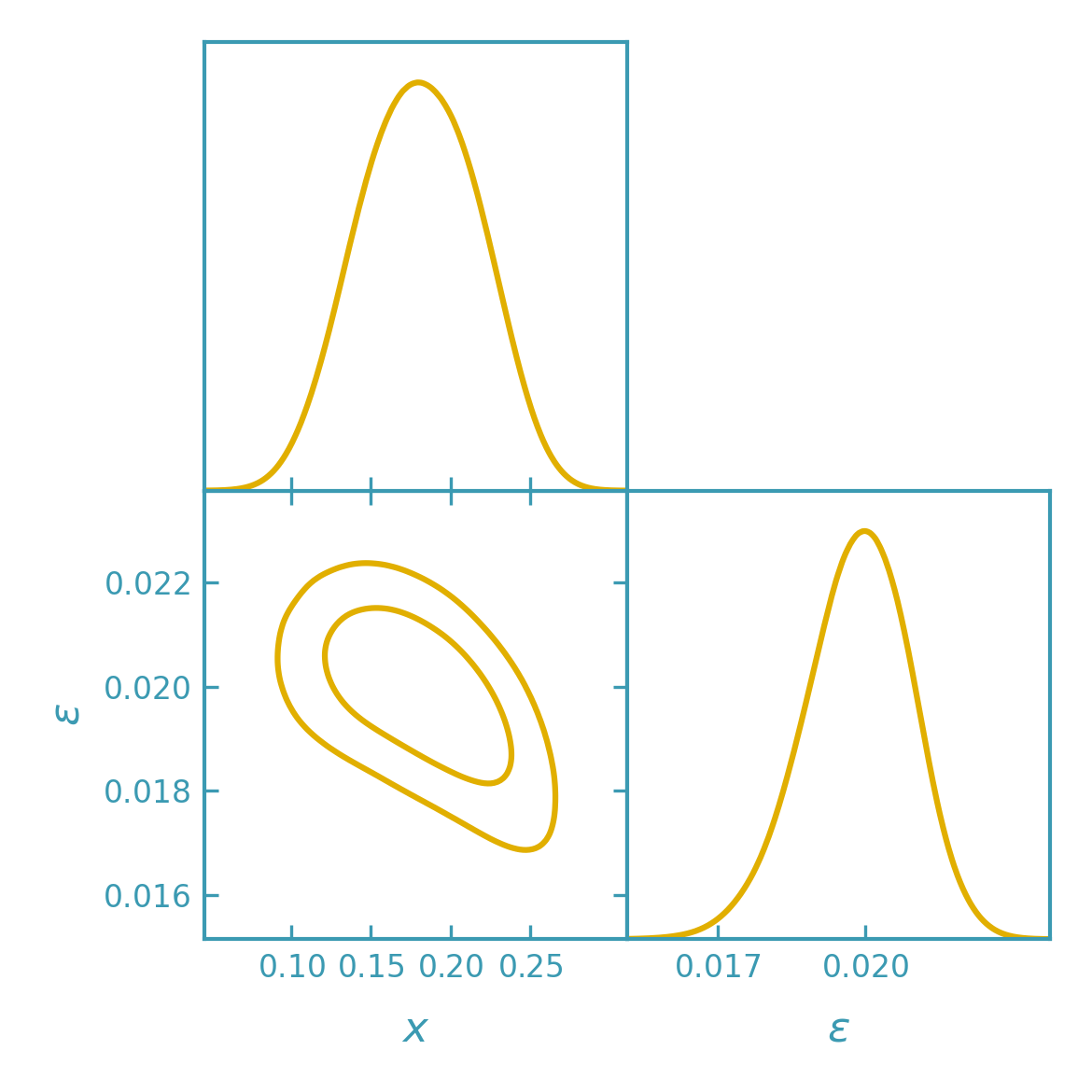
Sampling the DALI posterior with importance sampling#
>>> import numpy as np
>>> from getdist import plots as getdist_plots
>>> from derivkit import ForecastKit
>>> def model_2d(theta):
... # Nonlinear forward model with a curved parameter degeneracy
... # (informally referred to as a "banana"-shaped posterior).
... x, eps = float(theta[0]), float(theta[1])
... k = 3.0
... a = 4.0
... c = 6.0
... o1 = 1e2 * np.exp((x - k * eps) ** 2) * np.exp(a * eps)
... o2 = 4e1 * np.exp(0.5 * x) * (1.0 + 0.3 * eps + c * (eps**3))
... return np.array([o1, o2], dtype=float)
>>> theta0 = np.array([0.18, 0.02], dtype=float)
>>> cov = np.array([[1.0, 0.95],
... [0.95, 1.0]], dtype=float)
>>> fk = ForecastKit(function=model_2d, theta0=theta0, cov=cov)
>>> dali = fk.dali(forecast_order=2)
>>> samples = fk.getdist_dali_importance(
... dali=dali,
... names=["x", "eps"],
... labels=[r"x", r"\epsilon"],
... label="DALI (importance)",
... n_samples=80_000,
... seed=0,
... kernel_scale=1.3,
... )
>>> dk_yellow = "#e1af00"
>>> line_width = 1.5
>>> plotter = getdist_plots.get_subplot_plotter(width_inch=3.9)
>>> plotter.settings.linewidth_contour = line_width
>>> plotter.settings.linewidth = line_width
>>> plotter.settings.figure_legend_frame = False
>>> plotter.settings.legend_rect_border = False
>>> plotter.triangle_plot(
... [samples],
... params=["x", "eps"],
... filled=False,
... contour_colors=[dk_yellow],
... contour_lws=[line_width],
... contour_ls=["-"],
... )
>>> samples.numrows > 0
True
(png)
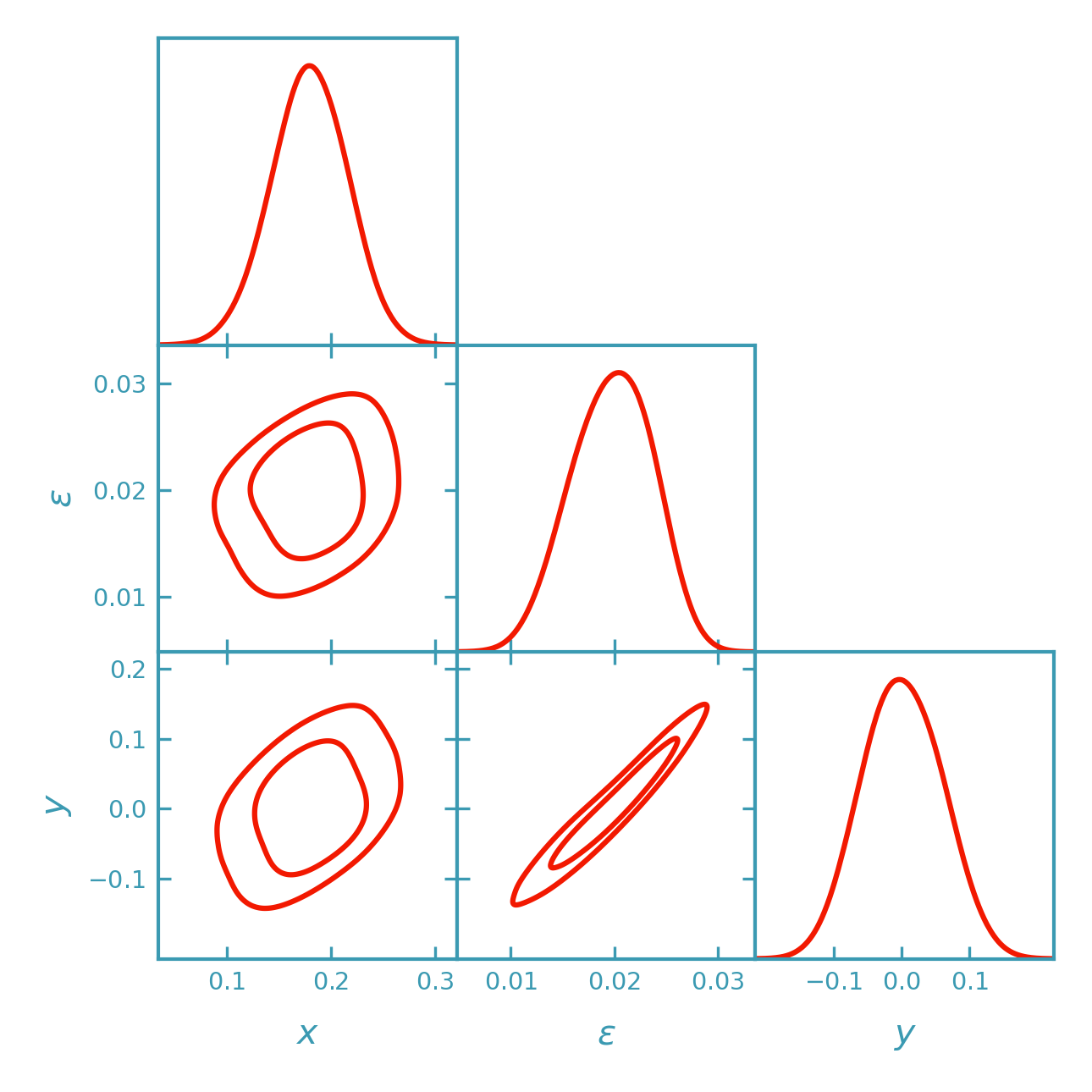
Three-parameter nonlinear example (emcee)#
This section extends the 2D example to three parameters
theta = [x, eps, y]. The forward model is constructed to produce a
nonlinear posterior with pronounced parameter degeneracies.
An additional coupling to y introduces further structure
while preserving the dominant nonlinear features.
o1uses(x - k*eps - q*y)^2to preserve the curved ridge in(x, eps)while introducing structure in(x, y)and(eps, y).o2adds a mild dependence onythrough an exponential prefactor.
>>> import numpy as np
>>> from getdist import plots as getdist_plots
>>> from derivkit import ForecastKit
>>> def model_3d(theta):
... # A nonlinear model with 3 parameters:
... x, eps, y = float(theta[0]), float(theta[1]), float(theta[2])
... k = 3.0
... q = 0.7
... a = 4.0
... c = 6.0
... r = 0.25
... o1 = 1e2 * np.exp((x - k * eps - q * y) ** 2) * np.exp(a * eps)
... o2 = 4e1 * np.exp(0.5 * (x + r * y)) * (1.0 + 0.3 * eps + c * (eps**3))
... return np.array([o1, o2], dtype=float)
>>> theta0 = np.array([0.18, 0.02, 0.00], dtype=float)
>>> cov = np.array([[1.0, 0.95],
... [0.95, 1.0]], dtype=float)
>>> prior_bounds = [(-0.4, 0.8), (-0.25, 0.25), (-0.4, 0.4)]
>>> fk = ForecastKit(function=model_3d, theta0=theta0, cov=cov)
>>> dali = fk.dali(forecast_order=2)
>>> samples = fk.getdist_dali_emcee(
... dali=dali,
... names=["x", "eps", "y"],
... labels=[r"x", r"\epsilon", r"y"],
... label="DALI (emcee, 3D)",
... prior_bounds=prior_bounds,
... )
>>> dk_red = "#f21901"
>>> dk_yellow = "#e1af00"
>>> dk_blue = "#3b9ab2"
>>> line_width = 1.5
>>> plotter = getdist_plots.get_subplot_plotter(width_inch=4.3)
>>> plotter.settings.linewidth_contour = line_width
>>> plotter.settings.linewidth = line_width
>>> plotter.settings.figure_legend_frame = False
>>> plotter.settings.legend_rect_border = False
>>> plotter.triangle_plot(
... [samples],
... params=["x", "eps", "y"],
... filled=False,
... contour_colors=[dk_red, dk_blue, dk_yellow],
... contour_lws=[line_width, line_width, line_width],
... contour_ls=["-", "-", "-"],
... )
>>> samples.numrows > 0
True
(png)
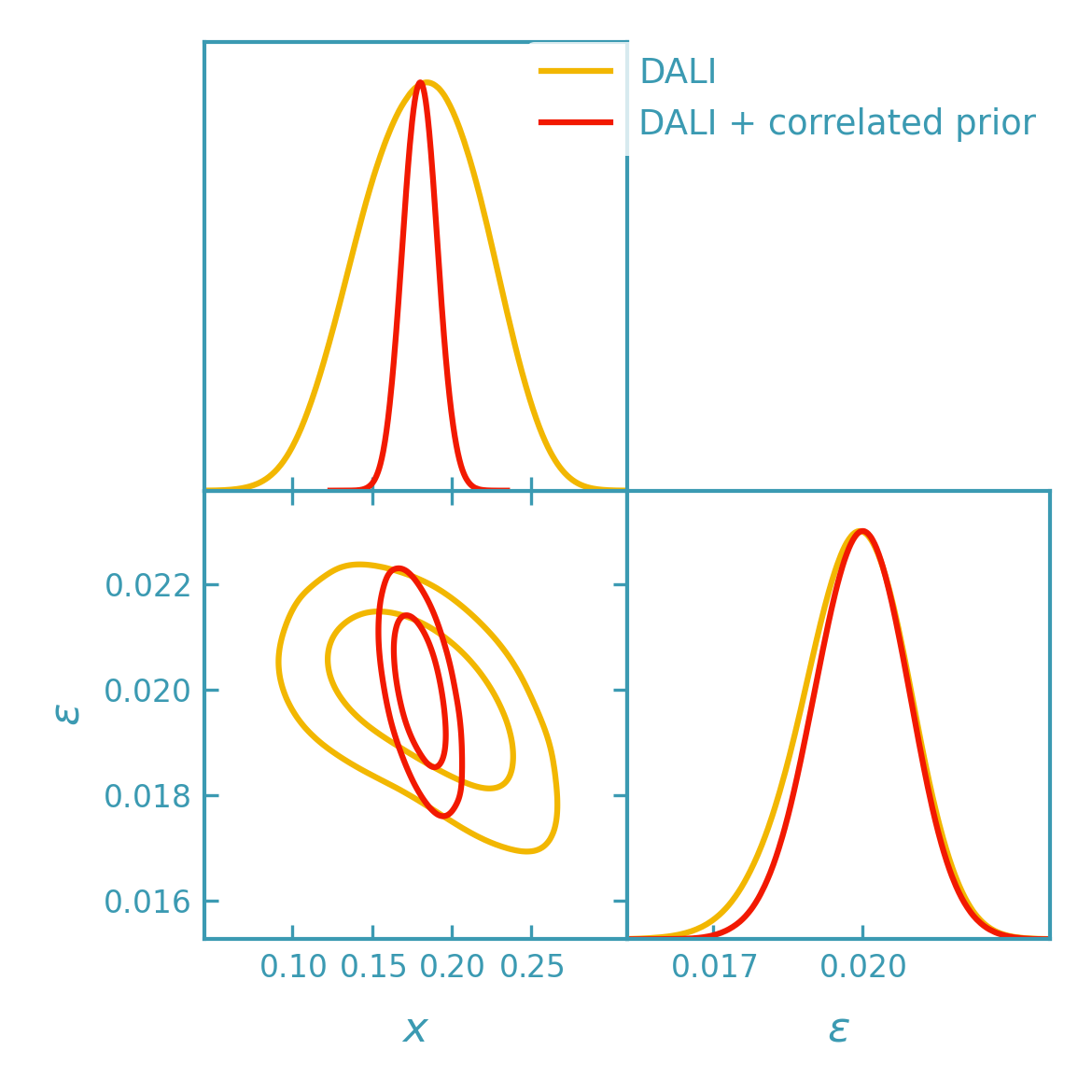
Including priors in DALI contours#
Priors can be included in DALI sampling by passing them directly to the
DerivKit GetDist helpers via prior_terms and/or prior_bounds.
These are evaluated as part of the DALI log-posterior during sampling.
Sampler bounds mainly truncate the sampled region, while informative priors (especially correlated multivariate priors) can change the shape and orientation of the contours.
>>> import numpy as np
>>> from getdist import plots as getdist_plots
>>> from derivkit import ForecastKit
>>> def model_2d(theta):
... x, eps = float(theta[0]), float(theta[1])
... k = 3.0
... a = 4.0
... c = 6.0
... o1 = 1e2 * np.exp((x - k * eps) ** 2) * np.exp(a * eps)
... o2 = 4e1 * np.exp(0.5 * x) * (1.0 + 0.3 * eps + c * (eps**3))
... return np.array([o1, o2], dtype=float)
>>> theta0 = np.array([0.18, 0.02], dtype=float)
>>> cov = np.array([[1.0, 0.95],
... [0.95, 1.0]], dtype=float)
>>> fk = ForecastKit(function=model_2d, theta0=theta0, cov=cov)
>>> dali = fk.dali(forecast_order=2)
>>> # Baseline: no priors
>>> samples_base = fk.getdist_dali_emcee(
... dali=dali,
... names=["x", "eps"],
... labels=[r"x", r"\epsilon"],
... label="DALI",
... )
>>> # With priors: wide bounds + a correlated multivariate Gaussian prior
>>> prior_bounds = [(-1.5, 1.5), (-0.8, 0.8)]
>>> # Strong correlated prior centered near theta0
>>> mu = np.array([0.18, 0.02], dtype=float)
>>> sx, seps, rho = 0.03, 0.006, -0.95
>>> cov_prior = np.array(
... [[sx * sx, rho * sx * seps],
... [rho * sx * seps, seps * seps]],
... dtype=float,
... )
>>> prior_terms = [("gaussian", {"mean": mu, "cov": cov_prior})]
>>> samples_prior = fk.getdist_dali_emcee(
... dali=dali,
... names=["x", "eps"],
... labels=[r"x", r"\epsilon"],
... label="DALI + correlated prior",
... prior_bounds=prior_bounds,
... prior_terms=prior_terms,
... )
>>> dk_red = "#f21901"
>>> dk_yellow = "#f2b701"
>>> line_width = 1.5
>>> plotter = getdist_plots.get_subplot_plotter(width_inch=3.9)
>>> plotter.settings.linewidth_contour = line_width
>>> plotter.settings.linewidth = line_width
>>> plotter.settings.figure_legend_frame = False
>>> plotter.settings.legend_rect_border = False
>>> plotter.triangle_plot(
... [samples_base, samples_prior],
... params=["x", "eps"],
... filled=[False, False],
... contour_colors=[dk_yellow, dk_red],
... contour_lws=[line_width, line_width],
... contour_ls=["-", "-"],
... )
>>> (samples_base.numrows > 0) and (samples_prior.numrows > 0)
True
(png)
Notes and conventions#
The non-Gaussianity here comes from the nonlinear forward model.
getdist.MCSamples.loglikesstores minus the log-posterior (up to an additive constant), following GetDist conventions.Importance sampling uses a Fisher–Gaussian proposal;
kernel_scalecontrols its width. If weights become extremely uneven, try increasing the scale slightly.Importance sampling is intended for fast visualization and exploratory work, and is reliable when the Fisher–Gaussian proposal closely matches the DALI posterior.
For science analyses requiring a robust exploration of the posterior, including non-Gaussian structure and tails, we recommend using emcee to sample the DALI-expanded posterior.
Typical workflow#
Compute DALI tensors with
ForecastKit(e.g.dali = fk.dali(forecast_order=2)).Use importance sampling for fast visualization and iteration.
Switch to
emceewhen robustness is required or strong non-Gaussianity leads to unstable importance weights.Visualize and compare results using GetDist triangle plots.
See also#
derivkit.forecast_kit.ForecastKitderivkit.forecast_kit.ForecastKit.dali()derivkit.forecast_kit.ForecastKit.logposterior_dali()derivkit.forecast_kit.ForecastKit.getdist_dali_emcee()derivkit.forecast_kit.ForecastKit.getdist_dali_importance()