 Fisher contours#
Fisher contours#
This page shows how to visualize Fisher-matrix forecasts using GetDist,
starting from a Fisher matrix computed with
derivkit.forecast_kit.ForecastKit.
The focus here is what to do next once you already have a Fisher matrix: how to turn it into confidence contours or samples for quick inspection, comparison, and plotting.
If you are looking for:
how the Fisher matrix is defined and interpreted, see ForecastKit
how to compute a Fisher matrix with DerivKit, see Fisher matrix
Two complementary visualization workflows are supported:
conversion of a Fisher matrix into an analytic Gaussian for GetDist
Monte Carlo samples drawn from the Fisher Gaussian, returned as
getdist.MCSamples
Both outputs can be passed directly to GetDist plotting utilities (e.g. triangle / corner plots).
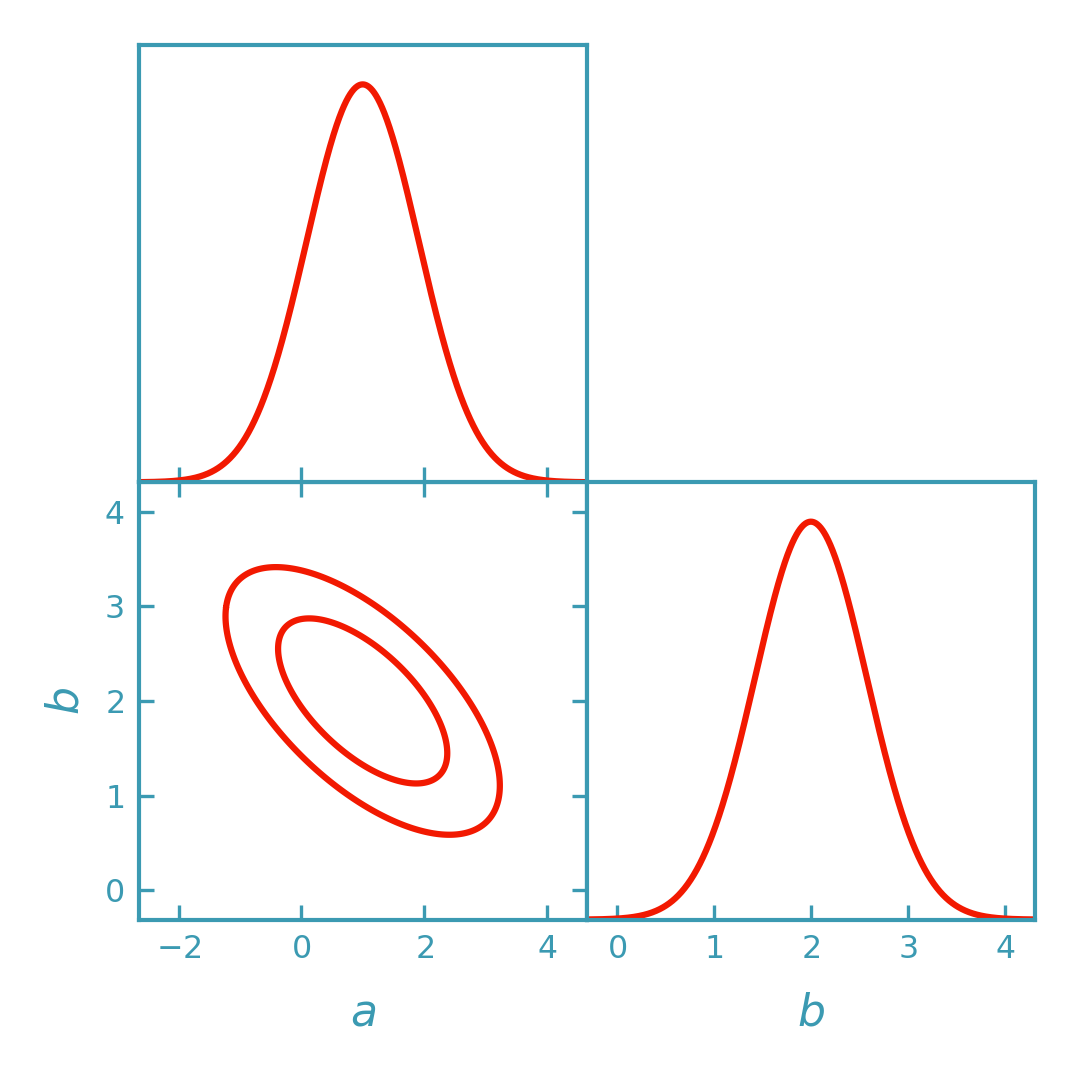
Analytic Gaussian (no sampling)#
Convert the Fisher matrix into an analytic Gaussian object compatible with GetDist, then plot Fisher ellipses using GetDist.
>>> import numpy as np
>>> from getdist import plots as getdist_plots
>>> from derivkit import ForecastKit
>>> # Define a simple toy model
>>> def model(theta):
... a, b = theta
... return np.array([a, b, a + 2.0 * b], dtype=float)
>>> # Fiducial parameters and covariance
>>> theta0 = np.array([1.0, 2.0])
>>> cov = np.eye(3)
>>> # Compute Fisher matrix
>>> fk = ForecastKit(function=model, theta0=theta0, cov=cov)
>>> fisher = fk.fisher(
... method="finite",
... stepsize=1e-2,
... num_points=5,
... extrapolation="ridders",
... levels=4,
... )
>>> # Convert Fisher matrix to analytic GetDist Gaussian
>>> gnd = fk.getdist_fisher_gaussian(
... fisher=fisher,
... names=["a", "b"],
... labels=[r"a", r"b"],
... label="Fisher (Gaussian)",
... )
>>> # Plot Fisher ellipses in DerivKit red (rendered by the docs build)
>>> dk_blue = "#3b9ab2"
>>> dk_red = "#f21901"
>>> line_width = 1.5
>>> plotter = getdist_plots.get_subplot_plotter(width_inch=3.6)
>>> plotter.settings.linewidth_contour = line_width
>>> plotter.settings.linewidth = line_width
>>> plotter.triangle_plot(
... [gnd],
... params=["a", "b"],
... filled=[False],
... contour_colors=[dk_red],
... contour_lws=[line_width],
... contour_ls=["-"],
... )
>>> isinstance(gnd, object)
True
(png)
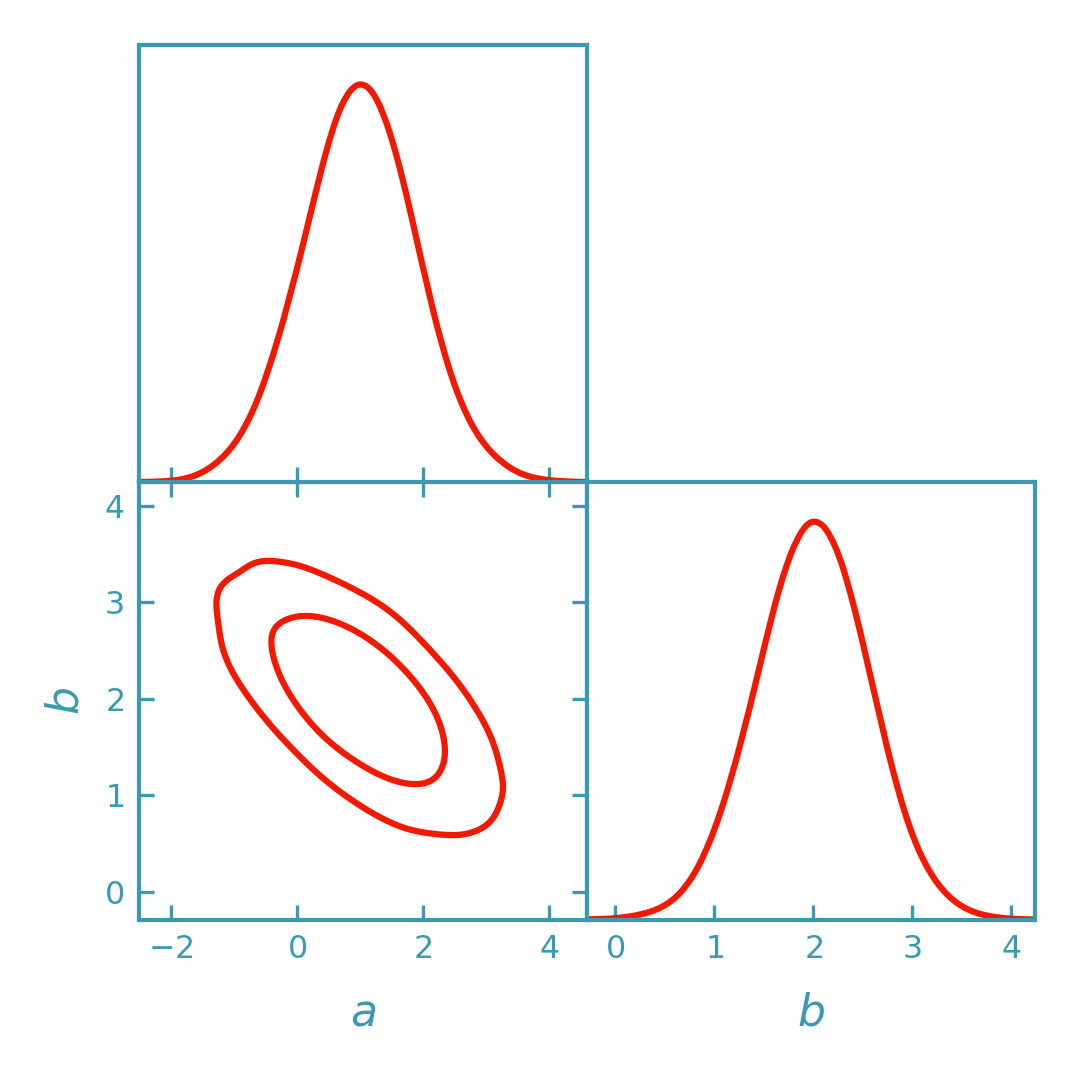
Sampling from the Fisher Gaussian#
For more flexibility (e.g. marginal histograms, bounds, or combining with other samples), draw Monte Carlo samples from the Fisher Gaussian and plot them with GetDist.
>>> import numpy as np
>>> from getdist import plots as getdist_plots
>>> from derivkit import ForecastKit
>>> # Define a simple toy model
>>> def model(theta):
... a, b = theta
... return np.array([a, b, a + 2.0 * b], dtype=float)
>>> # Fiducial parameters and covariance
>>> theta0 = np.array([1.0, 2.0])
>>> cov = np.eye(3)
>>> # Compute Fisher matrix
>>> fk = ForecastKit(function=model, theta0=theta0, cov=cov)
>>> fisher = fk.fisher(
... method="finite",
... stepsize=1e-2,
... num_points=5,
... extrapolation="ridders",
... levels=4,
... )
>>> # Draw samples from the Fisher Gaussian
>>> samples = fk.getdist_fisher_samples(
... fisher=fisher,
... names=["a", "b"],
... labels=[r"a", r"b"],
... store_loglikes=True,
... label="Fisher (samples)",
... )
>>> # Plot sample-based contours in DerivKit red (rendered by the docs build)
>>> dk_blue = "#3b9ab2"
>>> dk_red = "#f21901"
>>> line_width = 1.5
>>> plotter = getdist_plots.get_subplot_plotter(width_inch=3.6)
>>> plotter.settings.linewidth_contour = line_width
>>> plotter.settings.linewidth = line_width
>>> plotter.triangle_plot(
... samples,
... params=["a", "b"],
... filled=False,
... contour_colors=[dk_red],
... contour_lws=[line_width],
... contour_ls=["-"],
... )
>>> samples.numrows > 0
True
(png)
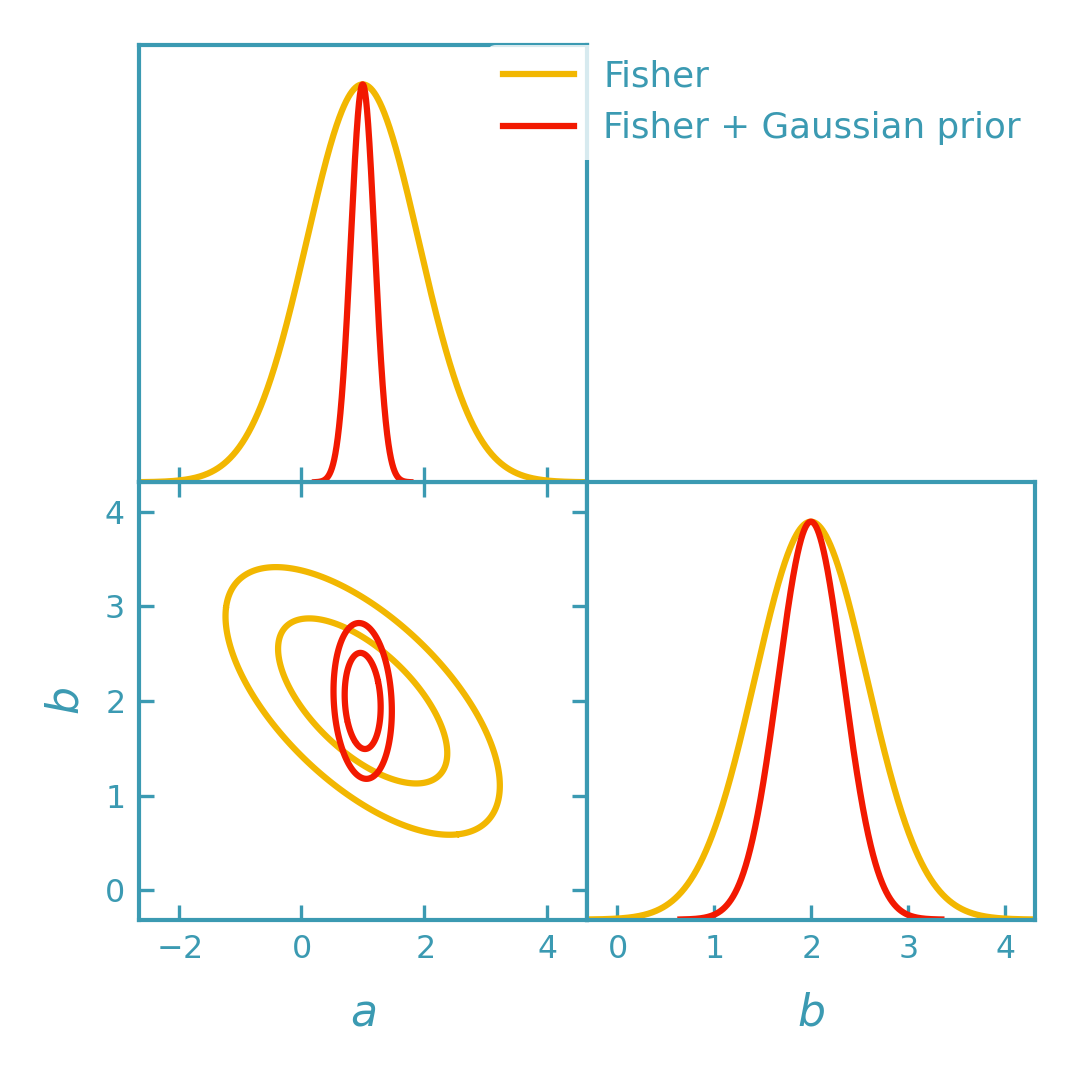
Including Gaussian priors in Fisher forecast#
Gaussian priors can be included by adding their precision matrix (the inverse prior covariance) to the Fisher matrix before converting to GetDist objects. Below we overlay the original Fisher contours (red) with the Fisher+prior contours (yellow).
>>> import numpy as np
>>> from getdist import plots as getdist_plots
>>> from derivkit import ForecastKit
>>> np.set_printoptions(precision=8, suppress=True)
>>> # Same toy model as above
>>> def model(theta):
... a, b = theta
... return np.array([a, b, a + 2.0 * b], dtype=float)
>>> theta0 = np.array([1.0, 2.0])
>>> cov = np.eye(3)
>>> # Fisher from the example above
>>> fk = ForecastKit(function=model, theta0=theta0, cov=cov)
>>> fisher_like = fk.fisher()
>>> # Gaussian prior: sigma_a = 0.2, sigma_b = 0.5 (diagonal prior covariance)
>>> sigma_prior = np.array([0.2, 0.5], dtype=float)
>>> fisher_prior = np.diag(1.0 / sigma_prior**2)
>>> fisher_post = fisher_like + fisher_prior
>>> # Convert both to analytic GetDist Gaussians
>>> g_like = fk.getdist_fisher_gaussian(
... fisher=fisher_like,
... names=["a", "b"],
... labels=[r"a", r"b"],
... label="Fisher",
... )
>>> g_post = fk.getdist_fisher_gaussian(
... fisher=fisher_post,
... names=["a", "b"],
... labels=[r"a", r"b"],
... label="Fisher + Gaussian prior",
... )
>>> # Overlay contours: red (likelihoods-only) and yellow (with prior)
>>> dk_red = "#f21901"
>>> dk_yellow = "#f2b701"
>>> line_width = 1.5
>>> plotter = getdist_plots.get_subplot_plotter(width_inch=3.6)
>>> plotter.settings.linewidth_contour = line_width
>>> plotter.settings.linewidth = line_width
>>> plotter.settings.figure_legend_frame = False
>>> plotter.settings.legend_rect_border = False
>>> plotter.triangle_plot(
... [g_like, g_post],
... params=["a", "b"],
... filled=[False, False],
... contour_colors=[dk_yellow, dk_red],
... contour_lws=[line_width, line_width],
... contour_ls=["-", "-"],
... )
>>> (g_like is not None) and (g_post is not None)
True
(png)
Notes and conventions#
The Fisher matrix is inverted using a pseudo-inverse to form the Gaussian covariance; regularization can be controlled via
rcond.getdist.MCSamples.loglikesstores minus the log-posterior (up to an additive constant), following GetDist conventions.Sampler bounds and priors are optional and intended for light truncation, not for defining complex posteriors.
Sampling-based Fisher contours are estimated via kernel density methods and may appear slightly irregular even for large sample sizes (e.g.
n_samples=100_000). This is expected and does not indicate an issue with the Fisher matrix itself.For strongly non-Gaussian posteriors or curved degeneracies, consider using the DALI expansion or a full sampler instead.
See also#
derivkit.forecast_kit.ForecastKitderivkit.forecast_kit.ForecastKit.fisher()derivkit.forecast_kit.ForecastKit.getdist_fisher_gaussian()derivkit.forecast_kit.ForecastKit.getdist_fisher_samples()